5 Standard errors
Appropriate standard errors depend on application-specific knowledge about how the data were generated. For example, appropriate standard errors are not the same for sampling variability in simple random samples, sampling variability in complex samples, and finite-sample inference with variation from random treatment assignment.
For the particular case of simple random samples, the package supports standard error estimation by the nonparametric bootstrap and 95% confidence intervals by a Normal approximation using the estimated standard error.
We first set up the environment
and simulate data
data <- pstratreg_sim(n = 100)and produce point estimates with a call to pstratreg.
pstratreg.out <- pstratreg(
formula_y = formula(y ~ x*a),
formula_s = formula(s ~ x*a),
data = data,
treatment_name = "a"
)To estimate standard errors and confidence intervals, hand the output of the call to pstratreg to the pstratreg_se function, optionally specifying the number r of bootstrap samples.
result_with_se <- pstratreg_se(pstratreg.out, r = 100)The output of a call to pstratreg_se is a data frame containing estimates and inferential quantities.
#> # A tibble: 2 × 5
#> estimand estimate se ci.min ci.max
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 effect_y_lower 0.0343 0.338 -0.627 0.696
#> 2 effect_y_upper 1.76 0.326 1.12 2.40See the end of this page for a glossary of estimand terms in the output.
5.1 Visualize the result
One way to use this output to create visualizations using ggplot. For example, the code below visualizes bounded effects on the outcome.
result_with_se %>%
filter(estimand %in% c("effect_y_lower","effect_y_naive","effect_y_upper")) %>%
mutate(estimand_label = case_when(
estimand == "effect_y_lower" ~ "(1)\nLower Bound",
estimand == "effect_y_naive" ~ "(2)\nEstimate if No\nPost-Treatment\nSelection",
estimand == "effect_y_upper" ~ "(3)\nUpper Bound"
)) %>%
ggplot(aes(x = estimand_label, y = estimate,
ymin = ci.min, ymax = ci.max)) +
geom_point() +
geom_errorbar() +
ylab("Average Causal Effect on Outcome\nAmong the Always-Valid") +
scale_x_discrete(name = "Bound Estimates")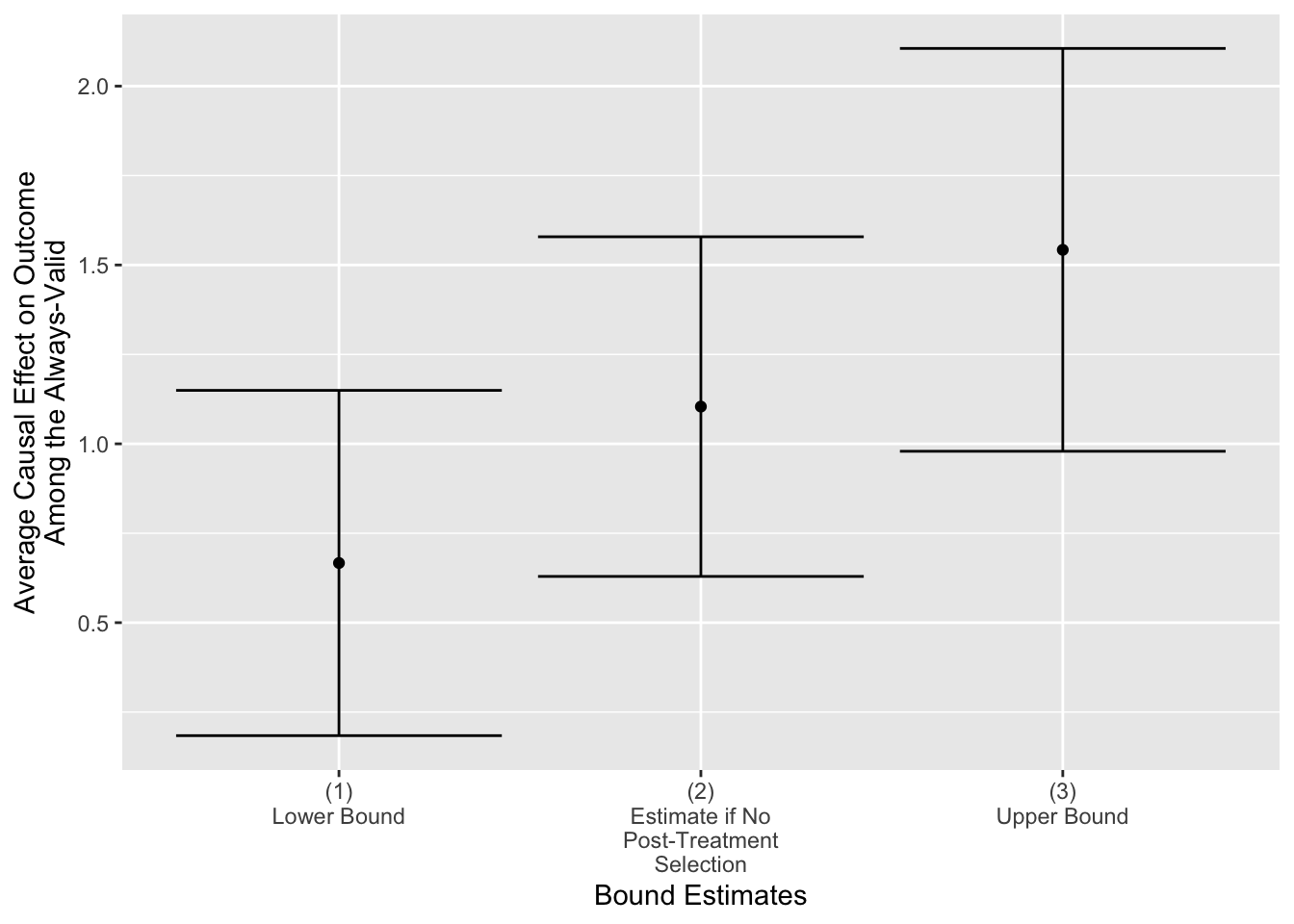
5.2 Glossary of estimands that result
Value of estimand
|
Definition | English |
|---|---|---|
effect_y_naive |
\(\hat{E}_{\text{Naive}}(Y^1-Y^0\mid S = \text{Always})\) | Average effect of treatment on outcome in always-valid stratum, estimated by assumption that potential outcomes are conditionally independent of \(M\) |
effect_y_lower |
\(\hat{E}_{\text{Lower}}(Y^1-Y^0\mid S = \text{Always})\) | Lower bound average effect of treatment on outcome in always-valid stratum |
effect_y_upper |
\(\hat{E}_{\text{Lower}}(Y^1-Y^0\mid S = \text{Always})\) | Upper bound average effect of treatment on outcome in always-valid stratum |
yhat_1_naive |
\(\hat{E}_{\text{Naive}}(Y^1\mid S = \text{Always})\) | Average outcome under treatment in the always-valid stratum, estimated by assumption potential outcomes are conditionally independent of \(M\) |
yhat_0_naive |
\(\hat{E}_{\text{Naive}}(Y^0\mid S = \text{Always})\) | Analogous to above, for outcome under control |
yhat_1_lower |
\(\hat{E}_{\text{Lower}}(Y^1\mid S = \text{Always})\) | Lower bound average outcome under treatment in the always-valid stratum |
yhat_0_lower |
\(\hat{E}_{\text{Lower}}(Y^0\mid S = \text{Always})\) | Analogous to above, for outcome under control |
yhat_1_upper |
\(\hat{E}_{\text{Upper}}(Y^1\mid S = \text{Always})\) | Upper bound average outcome under treatment in the always-valid stratum |
yhat_0_upper |
\(\hat{E}_{\text{Upper}}(Y^0\mid S = \text{Always})\) | Analogous to above, for outcome under control |
effect_m |
\(\hat{E}(M^1-M^0)\) | Average effect of treatment on mediator |
mhat1 |
\(\hat{E}(M^1)\) | Average mediator under treatment |
mhat1_trunc |
The above, truncated to comply with the user-defined positivity assumption | |
mhat0 |
\(\hat{E}(M^0)\) | Average mediator under control |
mhat0_trunc |
The above, truncated to comply with the user-defined positivity assumption |